THIS ARTICLE/PRESS RELEASE IS PAID FOR AND PRESENTED BY the University of OSLO - read more
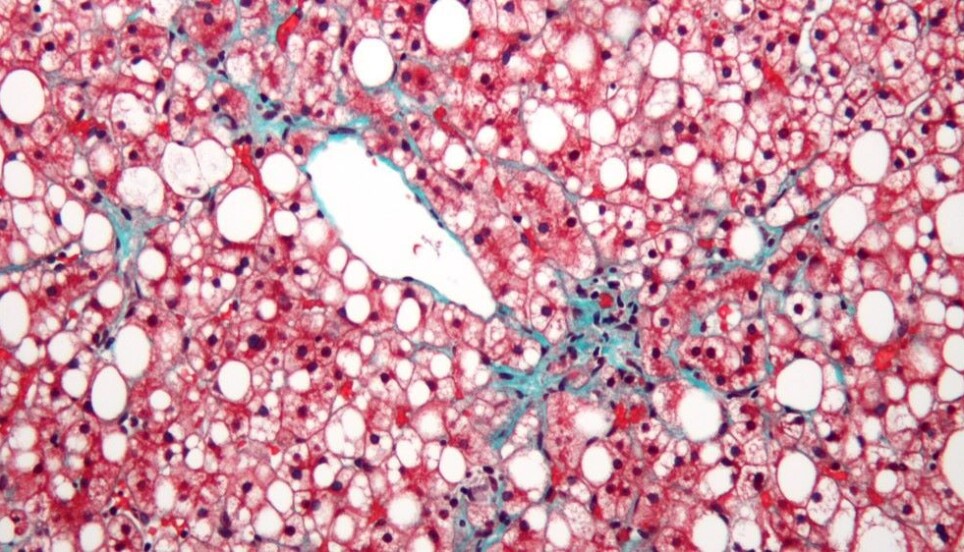
New genes discovered that regulate level of harmful fatty substances in liver
New Associate Professor at IBMS Frode Norheim identifies genes that could cause serious liver disease.
As many as one in every three Norwegians could have a fatty liver variant that is not caused by alcohol abuse. The disease is called non-alcoholic fatty liver disease, NAFLD, and is the most common liver disease in the Western world.

The disease is characterised by a build-up of fat in the liver. In some people, this fat accumulation may lead to inflammation and liver damage. We refer to such damage as non-alcoholic steatohepatitis. Over time, the disease can lead to cirrhosis or cancer. Some researchers estimate that half of all Americans will develop NAFLD by 2030.
The key is in the fatty substances
We have not yet identified all of the factors that contribute to some of us going from a build-up of fat in the liver to serious damage. But we have a great deal of evidence to show that our genes play a part. Researchers also think that certain lipids that are not stored correctly can lead to liver damage.
Previous research has largely focused on the total level of fat in the liver. What we do not yet know is how these lipids are regulated and whether they play a part in the cause of damage to the liver. Lipids is a common term for various fatty substances.
Study conducted by new Associate Professor at IBMS
A new study has shown which genes are important for controlling the amount of fatty substances in the liver and that could be important in the development of fatty liver disease and liver damage. The study was led by Frode Norheim, newly appointed Associate Professor at the Institute of Basic Medical Studies.
Two specific genes identified
The researchers wanted to identify the fatty substances that were most harmful to the liver. Damage occurs, among other things, as a result of patients becoming insulin-resistant and inflammation occurring in the liver.
“We examined several hundred fatty substances in the livers of hundreds of different strains of mice. This part of the research took place over three years at UCLA,” Norheim explains.
“One of the genes we studied in detail proved to be important for obesity and the storage of fat, known as triglycerides, in mice. This gene is called Map2k6,” he explains.
Another gene had an effect on the concentration of the fatty substance phosphatidylcholine in the liver. This gene is called Ifi203.
“Previous studies have shown that the balance between the two fatty substances is important in whether or not damage occurs in fatty livers,” Frode Norheim says.
The two genes were identified using so-called genome-wide association studies (GWAS).
These are studies that investigate the effect of genetic variants in a large number of people or, in this case, mice.
Potential future medicines
“Our hope for the future is that we can contribute to developing medicines that can regulate the level of harmful fatty substances. We could achieve this through a medicine that is effective in the two genes.”
The study was conducted by researcher Frode Norheim of the Institute of Basic Medical Sciences together with researchers at UCLA (University of California, Los Angeles). The study, which is a collaboration between Norheim and researchers at the University of California, Los Angeles (UCLA), was recently published in the journal Molecular Systems Biology.
Reference:
Frode Norheim et.al.: Genetic regulation of liver lipids in a mouse model of insulin resistance and hepatic steatosis, Molecular Systems Biology, 2021.
See more content from the University of Oslo:
-
Mainland Europe’s largest glacier may be halved by 2100
-
AI makes fake news more credible
-
What do our brains learn from surprises?
-
"A photograph is not automatically either true or false. It's a rhetorical device"
-
Queer opera singers: “I was too feminine, too ‘gay.’ I heard that on opera stages in both Asia and Europe”
-
Putin’s dream of the perfect family





































