This article was produced and financed by The Norwegian Veterinary Institute
Bacterial typing in the war against fish disease
Flavobacteria cause huge losses in fish farming worldwide. Researchers are now surveying the variation found amongst these bacteria to better control the disease.
Denne artikkelen er over ti år gammel og kan inneholde utdatert informasjon.
Flavobacteria are found in both fresh- and salt-water environments and cause significant problems in fish farming worldwide.
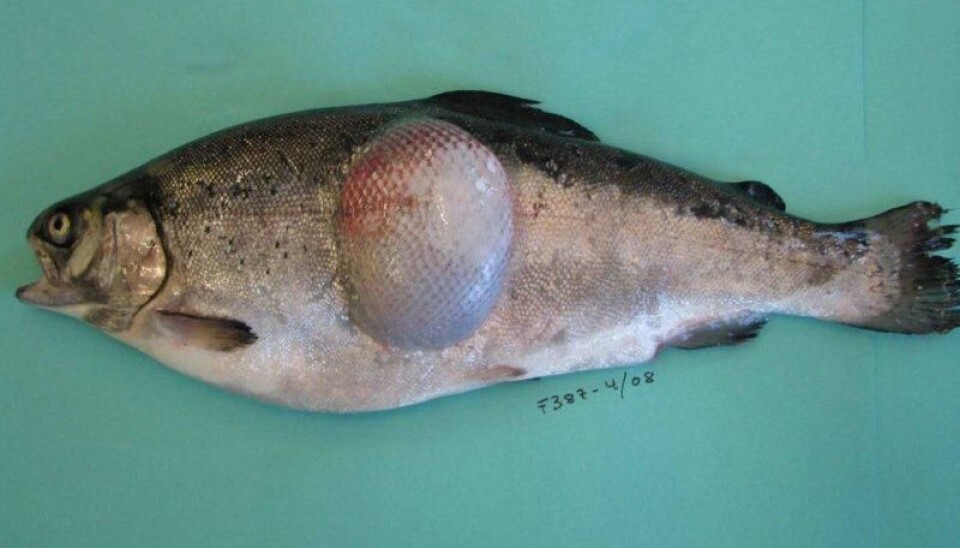
Although salmon may also be affected, Flavobacterium psychrophilum is responsible for one of the most significant diseases in rainbow trout in freshwater and can also cause disease in this fish species held in brackish water. Large losses of rainbow trout were registered in Norway during 2008.
The disease has occurred only sporadically in subsequent years, but still continues to pose a serious threat to Norwegian rainbow trout farming.
F. psychrophilum is highly infectious and results primarily in a systemic infection in rainbow trout with high associated mortalities.

In salmon, while infections are generally more associated with ulceration and ‘fin rot’, there is an increasing tendency towards systemic infection with significant mortality in juvenile salmon production sites. Several species of Tenacibaculum, a marine relative of Flavobacterium can also cause problems in sea-farmed fish at any stage in the production cycle.
European effort to control disease
An EU financed project entitled ‘Control of Flavobacteriaceae infections in European fish farms’ involving cooperative partners from France, Finland, Denmark, Switzerland, Italy and Norway, aims to develop tools to allow control of disease caused by Flavobacteria. This will be achieved by identification of the variation found amongst these bacteria and characterisation of the various types.
"Knowledge generated in this project will provide the basis for selection of vaccine candidates and development of diagnostic assays and vaccines. This will make detection and control possible e.g. through investigation of environmental samples and identification of apparently healthy ‘carrier’ fish," according to Hanne Nilsen, researcher at the Norwegian Veterinary Institute.
Currently such infections are normally treated using antibiotics.
Use of international standards allows comparison of data
"As several countries have agreed to use the same methods to characterise these bacteria, we can then compare and exchange information," says Nilsen.
"This is a huge advantage. We now have a database with over 1000 strains of F. psychrophilum from all over the world. Each individual strain of bacteria has been profiled using so-called MLST (multi-locus sequence typing) a technique which reveals differences in the genetic code. In this way we can form a global ‘picture’ of the disease caused by each bacterial variant. We can then identify which sequence types are most pathogenic, which have the greatest ability to spread, and which strains are currently ‘emerging’."
According to Nilsen, such studies will provide information on how the bacteria evolve with- and to- the host fish species and how they invade new countries.
Strain characterisation is an important tool for the aquaculture industry as it separates dangerous- from less dangerous- strains. Early strain-specific diagnosis is important as it allows use of an informed control strategy. This will reduce costs and increase fish welfare.
Genetic characterization
"In the first part of the project we have characterised Norwegian strains isolated from diseased fish. Bacterial genes coding for important ‘survival functions’ are selected for MLST," says the researcher.
The sequence of these genes can vary slightly between closely related bacteria, and the differences can be used to classify the bacterial isolates into different ‘groups’. The bacteria selected originate from different fish species with disease of varying severity.
"We have investigated bacterial strains from as many fish species and environments as possible, both from freshwater and brackish water, commercial and re-stocking hatcheries, fish from rivers and from the environment around fish farms. Samples were taken from routine investigations of diseased fish and from the biobank of the Norwegian Veterinary Institute. An equivalent study has been performed on Tenacibaculum bacteria isolated from diseased fish farmed in the sea."
The project has generated a huge amount of data which is now in the final stages of analysis. The information is collated in a global database in France hosted by the projects coordinating laboratory. Strains from each country are selected for examination of their ability to cause disease and form biofilm.
In addition the whole genome of selected isolates will be sequenced.
Rapid diagnostics allow better control
Important goals of the project are development of vaccines and more sensitive, specific and rapid diagnostic tools for identification of Flavobacteria.
Nilsen explains that through development of tools and methods which allow ‘screening’ of the environment and early identification of disease and carrier fish, the farmer will be able to make informed management decisions. The information can also be used to evaluate the risk of outbreak, or establish how infected a population of fish is.
"We believe the bacteria can be transmitted vertically, which means that the eggs can be infected. Even if disinfection is used, it can be difficult to eradicate the bacteria from eggs. We hope that through development of better diagnostic tools we can identify those fish which pose the highest threat of transmission to their offspring. By removal of these fish it may be possible to avoid or reduce the effect of large scale outbreaks," concludes Nilsen.

































