THIS ARTICLE/PRESS RELEASE IS PAID FOR AND PRESENTED BY NTNU Norwegian University of Science and Technology - read more

A new way to get cells to spill their secrets
Countless potentially useful enzymes are hidden all around us. NTNU researchers have developed a new method that could help us find them.
The natural world is a treasure trove of potentially useful enzymes, hidden in microorganisms living all around us. But finding them is tricky. A team of researchers at NTNU have developed a new method to break open cells that could help in the quest.
Enzymes are biological catalysts that can make industrial processes cheaper, less toxic, and more sustainable – and researchers are discovering new ones all the time. For example, a bacteria that uses two enzymes to break down plastic was found outside a plastic bottle recycling facility by Japanese researchers in 2016.
“Nature has a vast capacity for producing enzymes,” says Rahmi Lale, a synthetic biologist at NTNU’s Department of Biotechnology and Food Science.
Now, Lale and his colleagues have developed a new technique to break open cells in a controlled way – a vital step in screening for helpful enzymes. Crucially, the method leaves some cells intact, allowing them to be recovered and investigated further. The work, partly funded by the Research Council of Norway and the EU HORIZON 2020 programme, has been published in the journal ACS Synthetic Biology.
Millions of different cells
To find a new enzyme in nature, the first step is to identify an environment where microbes that need such an enzyme might live. Heat resistance is likely to be prized in the desert, for instance, whereas carbohydrate-degrading enzymes are more likely to be found in the human gut.
Next, researchers sift through material from that environment, such as a soil sample, to find genetic material. After isolating this environmental DNA, researchers cut it up into smaller pieces in the lab and paste it into well-studied microorganisms like E. coli (a process known as cloning) to screen for enzymatic reactions.
This leads to countless microbes, each of which could contain what the researchers are looking for. From this point the search becomes a 'needle in a haystack problem,' says Lale.
“It’s easy to collect DNA, it’s easy to clone them, and it’s easy to introduce them into these microorganisms,” he says. “But all of a sudden you have hundreds of millions of different cells. Every one of them carries something unique, but you don’t know which carry things that you are interested in.”
Screening the candidates
Using microfluidics – liquids flowing through tiny channels etched into a chip – allows researchers to screen the microbes 10,000 times faster than previous methods.
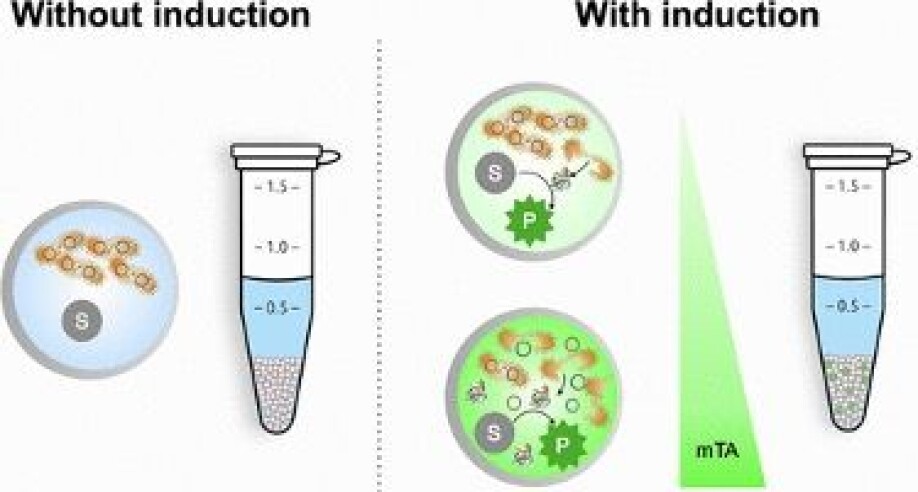
The microbes are contained in water droplets that sit in an oil-based carrier fluid. But because most of the potentially useful enzymes are made inside those microbes, the cells have to be opened up in order to test them.
To do this, Lale and his colleagues have developed a system that deliberately punctures the membrane of a cell, so its insides leak out into its surroundings. There, it can be tested for the presence of whatever kind of enzyme the team are looking for.
“We have a substrate that waits for the enzyme to come and interact with it,” says Lale. If there’s a positive match, it will trigger a reaction.
Controlling the holes
The standard way to break down the membrane of a cell, a process known as lysis, involves a chemical solution that is all but guaranteed to kill every cell in the droplet. But this presents a problem for researchers who, after finding an enzyme of interest, want to probe the microbe it came from further.
In contrast to chemical lysis, the “lysis-on-demand” system can be controlled by adjusting the concentration of the substance that induces lysis, meaning the researchers can deliberately leave some cells intact so they can be recovered later on.
“Because we can control how many holes we’re introducing, we can also control how many cells die,” says Lale. “We’re not killing them all, and that’s important.”
“If something of interest happens in one particular droplet, then we can recover that droplet,” he says. “Thanks to cells growing so quickly, we can take the droplet, put it in a growth medium and the next day have a billion cells again. Then recovery of DNA becomes a really simple task.”
The researchers confirmed their new technique in microfluidic chips they made in NTNU’s NanoLab.
From northern Norway to a UK compost heap
As well as boosting the search for useful molecules in nature, the technique could give a helping hand to researchers attempting to steer microbes towards making enzymes with particular traits.
By introducing genetic mutations in the lab, researchers can nudge the microbe in numerous different directions – then use microfluidic screening combined with the lysis-on-demand system to find the microbe that was nudged closer to making the kind of enzyme they want.
Lale and his colleagues have applied for a grant to explore this aspect of the research further, as part of a consortium led by colleagues at the University of Cambridge, UK. The researchers plan to use the system to sift through environmental DNA samples from various locations that could yield interesting enzymes, including the cold of northern Norway, the hot climate of southern Spain, and a UK compost heap.
“If you look into these interesting environments, the chances of finding something is higher,” says Lale.
Reference:
Che Fai Alex Wong et.al.: A Titratable Cell Lysis-on-Demand System for Droplet-Compartmentalized Ultrahigh-Throughput Screening in Functional Metagenomics and Directed Evolution. ACS Synth. Biol., 2021.
See more content from NTNU:
-
Fish farming is least harmful to the seabed in the north
-
Study: Centralising hospitals has reduced birth mortality
-
Early testing of schoolchildren: “We found absolutely no effect”
-
This determines whether your income level rises or falls
-
Why is nothing being done about the destruction of nature?“We hand over the data, but then it stops there"
-
Researchers now know more about why quick clay is so unstable





































